Updates | Technology
Publications
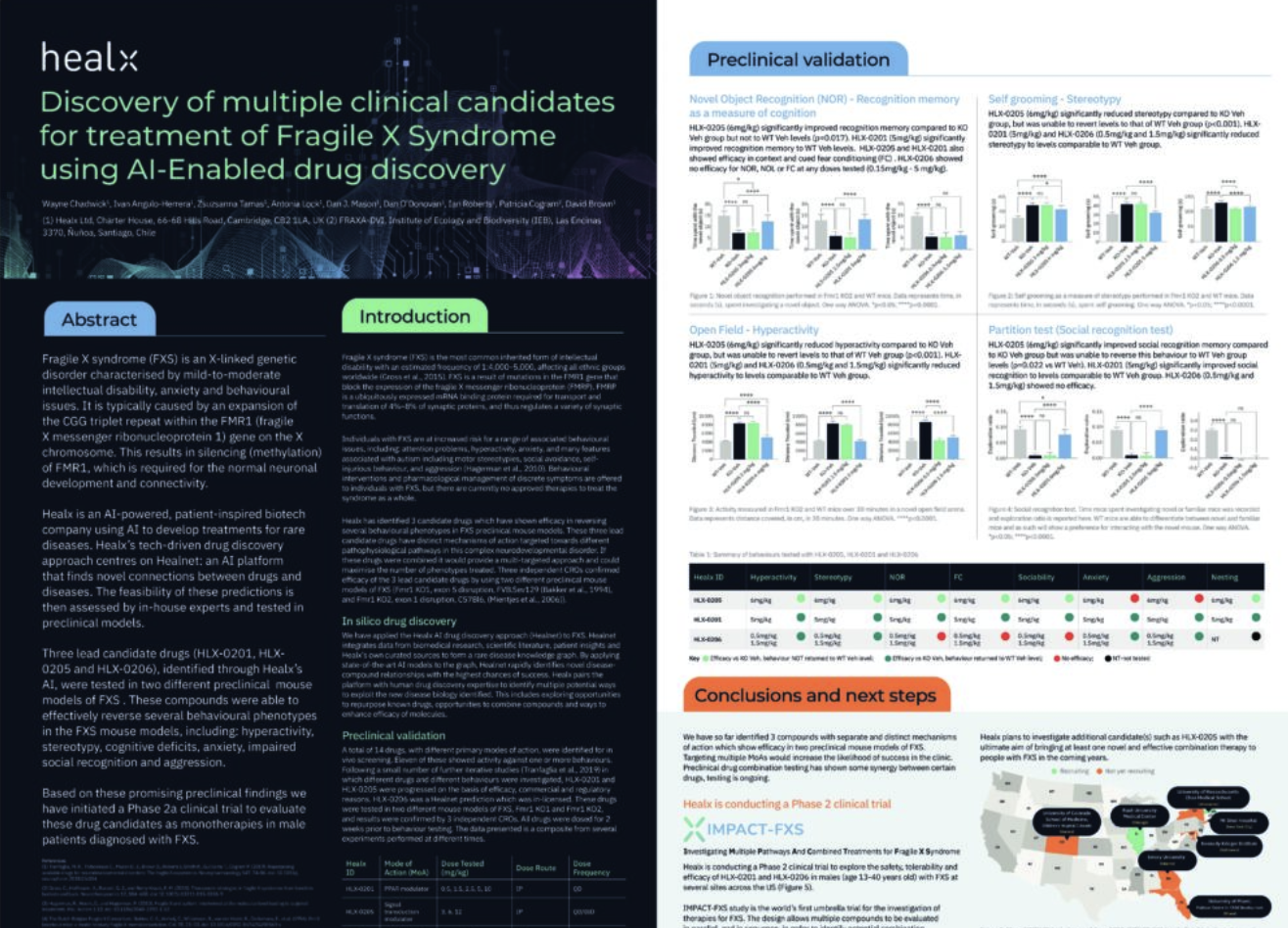
Fragile X syndrome (FXS) is an X-linked genetic disorder characterised by mild-to-moderate intellectual disability, anxiety and behavioural issues. It is typically caused by an expansion of the CGG triplet repeat within the FMR1 (fragile
X messenger ribonucleoprotein 1) gene on the X chromosome. This results in silencing (methylation) of FMR1, which is required for the normal neuronal development and connectivity.

Information Extraction (IE) from text refers to the task of extracting structured knowledge from unstructured text. The task typically consists of a series of sub-tasks such as Named Entity Recognition and Relation Extraction. Sourcing entity and relation type specific training data is a major bottleneck in domains with limited resources such as biomedicine. In this work we present a slot filling approach to the task of biomedical IE, effectively replacing the need for entity and relation-specific training data, allowing us to deal with zero-shot settings. We follow the recently proposed paradigm of coupling a Tranformer-based bi-encoder, Dense Passage Retrieval, with a Transformer-based reading comprehension model to extract relations from biomedical text. We assemble a biomedical slot filling dataset for both retrieval and reading comprehension and conduct a series of experiments demonstrating that our approach outperforms a number of simpler baselines. We also evaluate our approach end-to-end for standard as well as zero-shot settings. Our work provides a fresh perspective on how to solve biomedical IE tasks, in the absence of relevant training data. Our code, models and datasets are available at this https URL.
Real-world Relation Extraction (RE) tasks are challenging to deal with, either due to limited training data or class imbalance issues. In this work, we present Data Augmented Relation Extraction (DARE), a simple method to augment training data by properly fine-tuning GPT-2 to generate examples for specific relation types. The generated training data is then used in combination with the gold dataset to train a BERT-based RE classifier. In a series of experiments we show the advantages of our method, which leads in improvements of up to 11 F1 score points against a strong baseline. Also, DARE achieves new state of the art in three widely used biomedical RE datasets surpassing the previous best results by 4.7 F1 points on average.
Combination effects of compounds are commonly discussed in relation to the treatment of complex and treatment resistance-prone illnesses like cancer, (auto-)immune disorders, and infectious diseases. However, traversing the whole combinatorial space experimentally is infeasible. Therefore, in the last decades, substantial efforts have been made in the field of (predictive) combination modeling. In this reference module, we will discuss the goals of drug combination modeling, the features and endpoints that are used to address combination effect, and the modeling strategies which have been developed. We conclude that data availability is the largest bottle neck in the combination modeling pipeline. Improved data management and availability is essential for drug combination modeling to provide treatment benefit.

Reasoning over paths in large scale knowledge graphs is an important problem for many applications. In this paper we discuss a simple approach to automatically build and rank paths between a source and target entity pair with learned embeddings using a knowledge base completion model (KBC). We assembled a knowledge graph by mining the available biomedical scientific literature and extracted a set of high frequency paths to use for validation. We demonstrate that our method is able to effectively rank a list of known paths between a pair of entities and also come up with plausible paths that are not present in the knowledge graph. For a given entity pair we are able to reconstruct the highest ranking path 60% of the time within the the top 10 ranked paths and achieve 49% mean average precision. Our approach is compositional since any KBC model that can produce vector representations of entities can be used.
We present a novel framework to deal with relation extraction tasks in cases where there is complete lack of supervision, either in the form of gold annotations, or relations from a knowledge base. Our approach leverages syntactic parsing and pre-trained word embeddings to extract few but precise relations,which are then used to annotate a larger cor-pus, in a manner identical to distant supervision. The resulting data set is employed to fine tune a pre-trained BERT model in order to perform relation extraction. Empirical evaluation on four data sets from the biomedical domain shows that our method significantly outperforms two simple baselines for unsupervised relation extraction and, even if not using any supervision at all, achieves slightly worse results than the state-of-the-art in three out of four data sets. Importantly, we show that it is possible to successfully fine tune a large pre-trained language model with noisy data, as op-posed to previous works that rely on gold data for fine tuning.

Many available drugs have been repurposed as treatments for neurodevelopmental disorders. In the specific case of fragile X syndrome, many clinical trials of available drugs have been conducted with the goal of disease modification. In some cases, detailed understanding of basic disease mechanisms has guided the choice of drugs for clinical trials, and several notable successes in fragile X clinical trials have led to common use of drugs such as minocycline in routine medical practice. Newer technologies like Disease-Gene Expression Matching (DGEM) may allow for more rapid identification of promising repurposing candidates. A DGEM study predicted that sulindac could be therapeutic for fragile X, and subsequent preclinical validation studies have shown promising results. The use of combinations of available drugs and nutraceuticals has the potential to greatly expand the options for repurposing, and may even be a viable business strategy.
This article is part of the Special Issue entitled ‘Drug Repurposing: old molecules, new ways to fast track drug discovery and development for CNS disorders’.
The parasite Plasmodium falciparum is the most lethal species of Plasmodium to cause serious malaria infection in humans, and with resistance developing rapidly novel treatment modalities are currently being sought, one of which being combinations of existing compounds. The discovery of combinations of antimalarial drugs that act synergistically with one another is hence of great importance; however an exhaustive experimental screen of large drug space in a pairwise manner is not an option. In this study we apply our machine learning approach, Combination Synergy Estimation (CoSynE), which can predict novel synergistic drug interactions using only prior

Given the high attrition rates, substantial costs and slow pace of new drug discovery and development, repurposing of 'old' drugs to treat both common and rare diseases is increasingly becoming an attractive proposition because it involves the use of de-risked compounds, with potentially lower overall development costs and shorter development timelines. Various data-driven and experimental approaches have been suggested for the identification of repurposable drug candidates; however, there are also major technological and regulatory challenges that need to be addressed. In this Review, we present approaches used for drug repurposing (also known as drug repositioning), discuss the challenges faced by the repurposing community and recommend innovative ways by which these challenges could be addressed to help realize the full potential of drug repurposing.
Combining disease relationships across multiple biological levels could aid our understanding of common processes taking place in disease, potentially indicating opportunities for drug sharing. Here, we propose a similarity fusion approach which accounts for differences in information content between different data types, allowing combination of each data type in a balanced manner.
Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5) gene cause a complex neurological disorder, characterized by infantile seizures, impairment of cognitive and motor skills and autistic features. Loss of Cdkl5 in mice affects dendritic spine maturation and dynamics but the underlying molecular mechanisms are still far from fully understood. Here we show that Cdkl5 deficiency in primary hippocampal neurons leads to deranged expression of the alpha-amino-3-hydroxy-5-methyl-4-iso-xazole propionic acid receptors (AMPA-R). In particular, a dramatic reduction of expression of the GluA2 subunit occurs concomitantly with its hyper-phosphorylation on Serine 880 and increased ubiquitination. Consequently, Cdkl5 silencing skews the composition of membrane-inserted AMPA-Rs towards the GluA2-lacking calcium-permeable form. Such derangement is likely to contribute, at least in part, to the altered synaptic functions and cognitive impairment linked to loss of Cdkl5. Importantly, we find that tianeptine, a cognitive enhancer and antidepressant drug, known to recruit and stabilise AMPA-Rs at the synaptic sites, can normalise the expression of membrane inserted AMPA-Rs as well as the number of PSD-95 clusters, suggesting its therapeutic potential for patients with mutations in CDKL5.
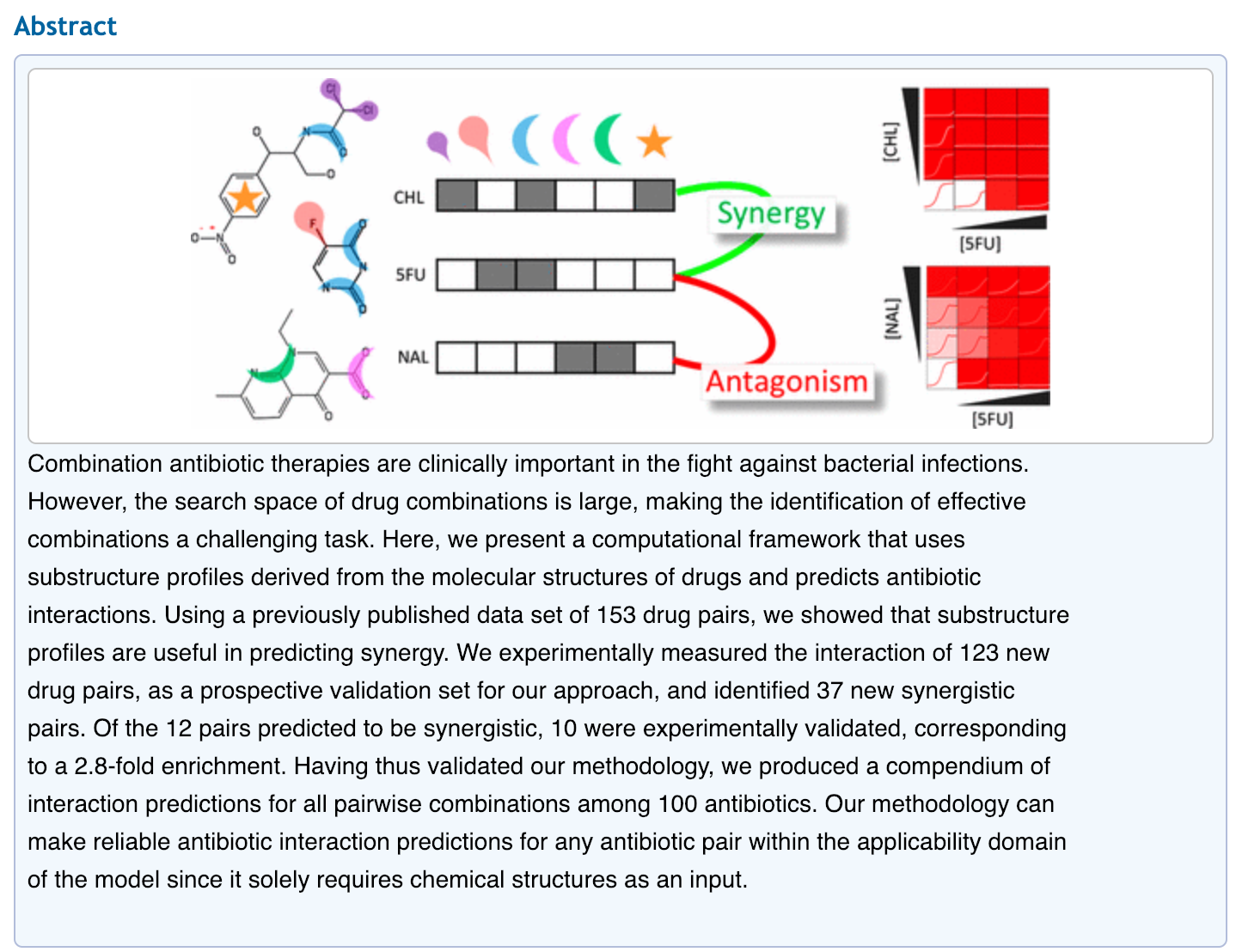
Combination antibiotic therapies are clinically important in the fight against bacterial infections. However, the search space of drug combinations is large, making the identification of effective combinations a challenging task. Here, we present a computational framework that uses substructure profiles derived from the molecular structures of drugs and predicts antibiotic interactions. Using a previously published data set of 153 drug pairs, we showed that substructure profiles are useful in predicting synergy. We experimentally measured the interaction of 123 new drug pairs, as a prospective validation set for our approach, and identified 37 new synergistic pairs. Of the 12 pairs predicted to be synergistic, 10 were experimentally validated, corresponding to a 2.8-fold enrichment. Having thus validated our methodology, we produced a compendium of interaction predictions for all pairwise combinations among 100 antibiotics. Our methodology can make reliable antibiotic interaction predictions for any antibiotic pair within the applicability domain of the model since it solely requires chemical structures as an input.

The development of treatments involving combinations of drugs is a promising approach towards combating complex or multifactorial disorders. However, the large number of compound combinations that can be generated, even from small compound collections, means that exhaustive experimental testing is infeasible. The ability to predict the behaviour of compound combinations in biological systems, whittling down the number of combinations to be tested, is therefore crucial. Here, we review the current state-of-the-art in the field of compound combination modelling, with the aim to support the development of approaches that, as we hope, will finally lead to an integration of chemical with systems-level biological information for predicting the effect of chemical mixtures.

Concern over antibiotic resistance is growing, and new classes of antibiotics, particularly against Gram-negative bacteria, are needed. However, even if the scientific hurdles can be overcome, it could take decades for sufficient numbers of such antibiotics to become available. As an interim solution, antibiotic resistance could be 'broken' by co-administering appropriate non-antibiotic drugs with failing antibiotics. Several marketed drugs that do not currently have antibacterial indications can either directly kill bacteria, reduce the antibiotic minimum inhibitory concentration when used in combination with existing antibiotics and/or modulate host defence through effects on host innate immunity, in particular by altering inflammation and autophagy. This article discusses how such 'antibiotic resistance breakers' could contribute to reducing the antibiotic resistance problem, and analyses a priority list of candidates for further investigation.
